DAS Workshop at the EBI
I made it to this years workshop about the Distributed Annotation System (DAS) at the European Bioinformatics Institute in Cambridge. Before we started working on openSNP I’ve never used the protocol but implementing one of the fields standards for sharing genetic information was definitely a thing we wanted to do with openSNP. We’ve worked on implementing this before the workshop but ran into some problems. After the first day of the workshop this already looks much better.
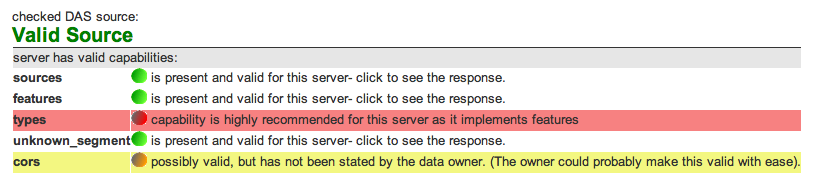
According to the validations performed by the DASRegistry we now pass the sources, features and unknown_segment-commands. And these are the only ones which are currently implemented in openSNP. This is a nice step forward. Today I’ll work on implementing the types-command as well, as this is mandatory for annotation-servers in the 1.6-specification of DAS (the 1.5[E]-specification doesn’t explicitly state the need for the types-command if you implement features). 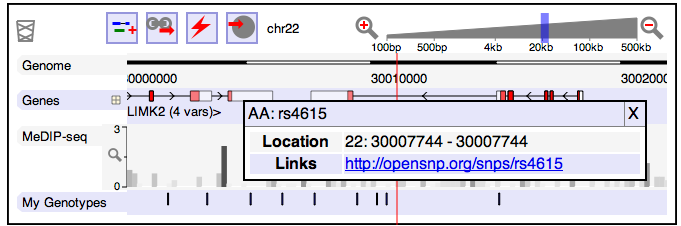
I also work on implementing some useful application for DAS right into openSNP: Biodalliance is a genome browser/viewer which runs right away in your browser using JavaScript and it can be used to easily embed genome views into web-pages. I’ll try to add the respective views of the genome right into the SNP-pages. The screenshot here is of the Biodalliance website and already includes my genotypes in the last track. So this should be straight forward to do.
